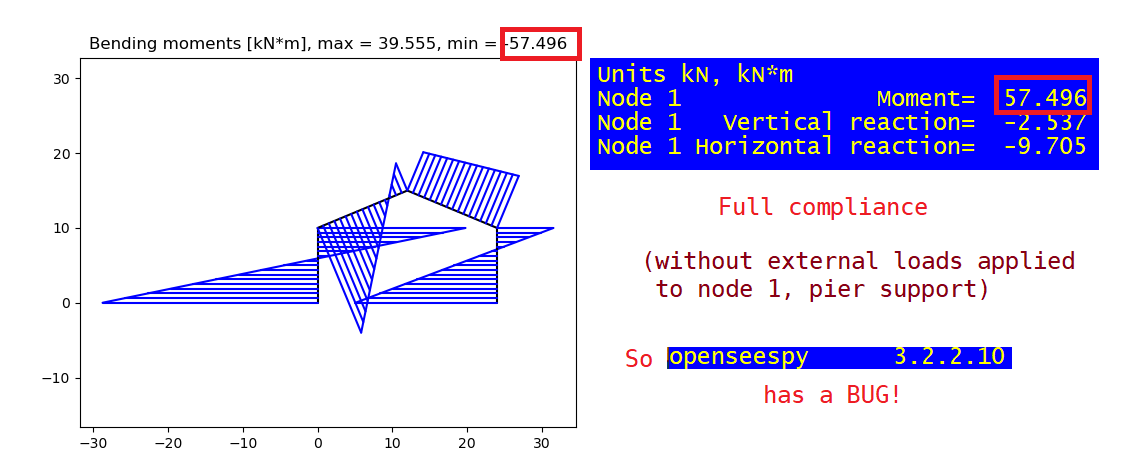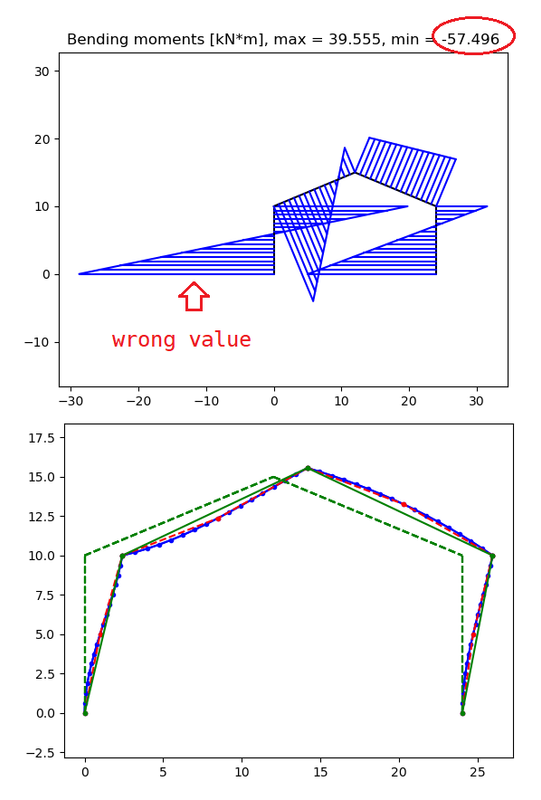
The 'strange matter' can be seen trough next schedule. Looking at bending moment on soil support (1) the proper value should be 69.996 (also confirmed by other sowtware) and not 57.496 as plotted in the diagram. Nevertheless, the one of node (5) is in perfect harmony, 37.874 kN*m.
Could it be a bug? Replies are welcomed. Cheers
Code: Select all
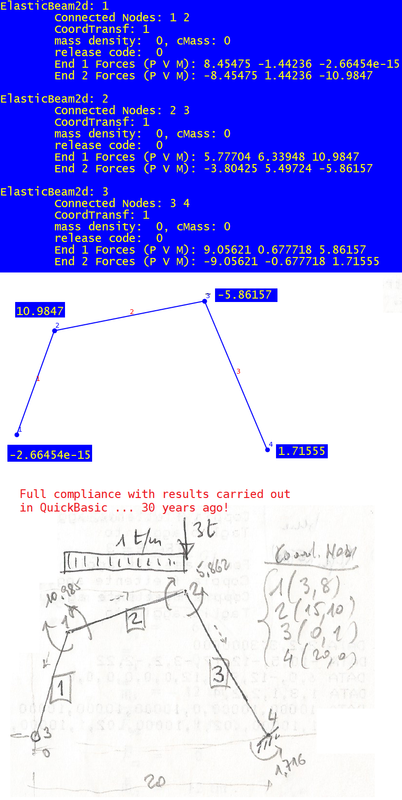
ElasticBeam2d: 1
Connected Nodes: 1 2
End 1 Forces (P V M): -2.53668 9.70513 57.4958 <<<<< discrepancy with reactions()
ElasticBeam2d: 4
End 2 Forces (P V M): -2.53668 -5.29487 37.8739 <<<<< in compliance with reactions()
-----------------------------------------
Node 1 Moment= 69.996
Node 1 Vertical reaction= -2.537
Node 1 Horizontal reaction= -17.205
Node 5 Moment= 37.874
Node 5 Vertical reaction= 2.537
Node 5 Horizontal reaction= -5.295
Code: Select all
# --- ese2_forum.py --- Jun.03, 2021 ----
#
import openseespy.opensees as ops
import openseespy.postprocessing.ops_vis as opsv
import matplotlib.pyplot as plt
import numpy as np
ops.wipe()
ops.model('basic', '-ndm', 2, '-ndf', 3)
# --- Gehler configuration ---
# A= 1000 stands for infinity, no deformations due to compressed areas
A = 1.e3
I = 1.
E = 1.
#
Ep = {1: [E, A, I],
2: [E, A, I],
3: [E, A, I],
4: [E, A, I]}
# Coord. nodes
ops.node(1, 0., 0.)
ops.node(2, 0., 10.)
ops.node(3, 12., 15.)
ops.node(4, 24., 10.)
ops.node(5, 24., 0.)
# Soil supports
ops.fix(1, 1, 1, 1) # fixed
ops.fix(5, 1, 1, 1) # fixed
ops.geomTransf('Linear',12)
# Elements (between nodes)
ops.element('elasticBeamColumn', 1, 1, 2, A, E, I, 12)
ops.element('elasticBeamColumn', 2, 2, 3, A, E, I, 12)
ops.element('elasticBeamColumn', 3, 3, 4, A, E, I, 12)
ops.element('elasticBeamColumn', 4, 4, 5, A, E, I, 12)
# External forces and couples
H1= 7.50; C1= -12.50
H2= 11.25; C2= 9.25
H3= 3.75; C3= 3.25
# --- Distributed loads ---
Wx= 0.
Wy= 0.
Ew = {3: ['-beamUniform', Wx, Wy]} # fictitious null load on beam 3
ops.timeSeries('Constant', 23)
ops.pattern('Plain', 1, 23)
# concentrated loads and couples applied
ops.load(1, H1, 0., C1)
ops.load(2, H2, 0., C2)
ops.load(3, H3, 0., C3)
for etag in Ew:
ops.eleLoad('-ele', etag, '-type', Ew[etag][0], Ew[etag][1],
Ew[etag][2])
ops.constraints('Transformation')
ops.numberer('RCM')
ops.system('BandGeneral')
ops.test('NormDispIncr', 1.0e-6, 6, 2)
ops.algorithm('Linear')
ops.integrator('LoadControl', 1)
ops.analysis('Static')
ops.analyze(1)
ops.printModel()
ops.reactions()
# plot model with tag labels
szer, wys = 16., 10.
fig = plt.figure(figsize=(szer/2.54, wys/2.54))
fig.subplots_adjust(left=.08, bottom=.08, right=.985, top=.94)
ax1 = plt.subplot(111)
opsv.plot_model()
sfacN, sfacV, sfacM = 5.e-1, 5.e-1, 5.e-1
# plot deformed model
plt.figure()
# plot_defo with optional arguments
sfac = opsv.plot_defo() #
print(f'sfac: {sfac}') # return sfac if automatically calculated
opsv.plot_defo(sfac, fmt_interp='b.-')
opsv.plot_defo(sfac, 5, interpFlag=0, fmt_nodes='bo-')
opsv.plot_defo(sfac, 3, endDispFlag=0, fmt_interp='r.--')
opsv.plot_defo(sfac, 2, fmt_interp='g.-')
# plot N, V, M forces diagrams
#
plt.figure()
minVal, maxVal = opsv.section_force_diagram_2d('N', Ew, sfacN)
plt.title(f'Axial forces [kN], max = {maxVal:.3f}, min = {minVal:.3f}')
plt.figure()
minVal, maxVal = opsv.section_force_diagram_2d('T', Ew, sfacV)
plt.title(f'Shear forces [kN], max = {maxVal:.3f}, min = {minVal:.3f}')
plt.figure()
minVal, maxVal = opsv.section_force_diagram_2d('M', Ew, sfacM)
plt.title(f'Bending moments [kN*m], max = {maxVal:.3f}, min = {minVal:.3f}')
# ----------
plt.show()
# ----------
print('\n Units kN, kN*m')
print(' Node 1 Moment= {:>7.3f}'. format (ops.nodeReaction(1,3)))
print(' Node 1 Vertical reaction= {:>7.3f}'. format (ops.nodeReaction(1,2)))
print(' Node 1 Horizontal reaction= {:>7.3f}'. format (ops.nodeReaction(1,1)))
print('\n')
print(' Node 5 Moment= {:>7.3f}'. format (ops.nodeReaction(5,3)))
print(' Node 5 Vertical reaction= {:>7.3f}'. format (ops.nodeReaction(5,2)))
print(' Node 5 Horizontal reaction= {:>7.3f}'. format (ops.nodeReaction(5,1)))
exit()
# EOF: ese2_forum.py
'''
C:\Training>python ese2_forum.py
Current Domain Information
Current Time: 1
tCommitted Time: 1
NODE DATA: NumNodes: 5
numComponents: 5
Node: 1
Coordinates : 0 0
Disps: 0 0 0
unbalanced Load: 7.5 0 -12.5
ID : -1 -1 -1
Node: 2
Coordinates : 0 10
Disps: 1257.27 0.0253668 -89.7014
unbalanced Load: 11.25 0 9.25
ID : 6 7 8
Node: 3
Coordinates : 12 15
Disps: 1134.29 295.157 56.2005
unbalanced Load: 3.75 0 3.25
ID : 3 4 5
Node: 4
Coordinates : 24 10
Disps: 1011.21 -0.0253668 -113.995
unbalanced Load: 0 0 0
ID : 0 1 2
Node: 5
Coordinates : 24 0
Disps: 0 0 0
unbalanced Load: 0 0 0
ID : -1 -1 -1
ELEMENT DATA: NumEle: 4
numComponents: 4
ElasticBeam2d: 1
Connected Nodes: 1 2
CoordTransf: 12
mass density: 0, cMass: 0
release code: 0
End 1 Forces (P V M): -2.53668 9.70513 57.4958
End 2 Forces (P V M): 2.53668 -9.70513 39.5555
ElasticBeam2d: 2
Connected Nodes: 2 3
CoordTransf: 12
mass density: 0, cMass: 0
release code: 0
End 1 Forces (P V M): 0.45039 -2.93573 -30.3055
End 2 Forces (P V M): -0.45039 2.93573 -7.85905
ElasticBeam2d: 3
Connected Nodes: 3 4
CoordTransf: 12
mass density: 0, cMass: 0
release code: 0
End 1 Forces (P V M): 5.86322 -0.305063 11.1091
End 2 Forces (P V M): -5.86322 0.305063 -15.0749
ElasticBeam2d: 4
Connected Nodes: 4 5
CoordTransf: 12
mass density: 0, cMass: 0
release code: 0
End 1 Forces (P V M): 2.53668 5.29487 15.0749
End 2 Forces (P V M): -2.53668 -5.29487 37.8739
SP_Constraints: numConstraints: 6
numComponents: 6SP_Constraint: 0 Node: 1 DOF: 1 ref value: 0 current value: 0
SP_Constraint: 1 Node: 1 DOF: 2 ref value: 0 current value: 0
SP_Constraint: 2 Node: 1 DOF: 3 ref value: 0 current value: 0
SP_Constraint: 3 Node: 5 DOF: 1 ref value: 0 current value: 0
SP_Constraint: 4 Node: 5 DOF: 2 ref value: 0 current value: 0
SP_Constraint: 5 Node: 5 DOF: 3 ref value: 0 current value: 0
Pressure_Constraints: numConstraints: 0
numComponents: 0
MP_Constraints: numConstraints: 0
numComponents: 0
LOAD PATTERNS: numPatterns: 1
numComponents: 1Load Pattern: 1
Scale Factor: 1
Constant Series: factor: 1
Nodal Loads:
numComponents: 3Nodal Load: 1 load : 7.5 0 -12.5
Nodal Load: 2 load : 11.25 0 9.25
Nodal Load: 3 load : 3.75 0 3.25
Elemental Loads:
numComponents: 1Beam2dUniformLoad - tag 0
Transverse: 0
Axial: 0
Element acted on: 3
Single Point Constraints:
numComponents: 0
PARAMETERS: numParameters: 0
numComponents: sfac: 0.0019089017319445234
Units kN, kN*m
Node 1 Moment= 69.996
Node 1 Vertical reaction= -2.537
Node 1 Horizontal reaction= -17.205
Node 5 Moment= 37.874
Node 5 Vertical reaction= 2.537
Node 5 Horizontal reaction= -5.295
'''